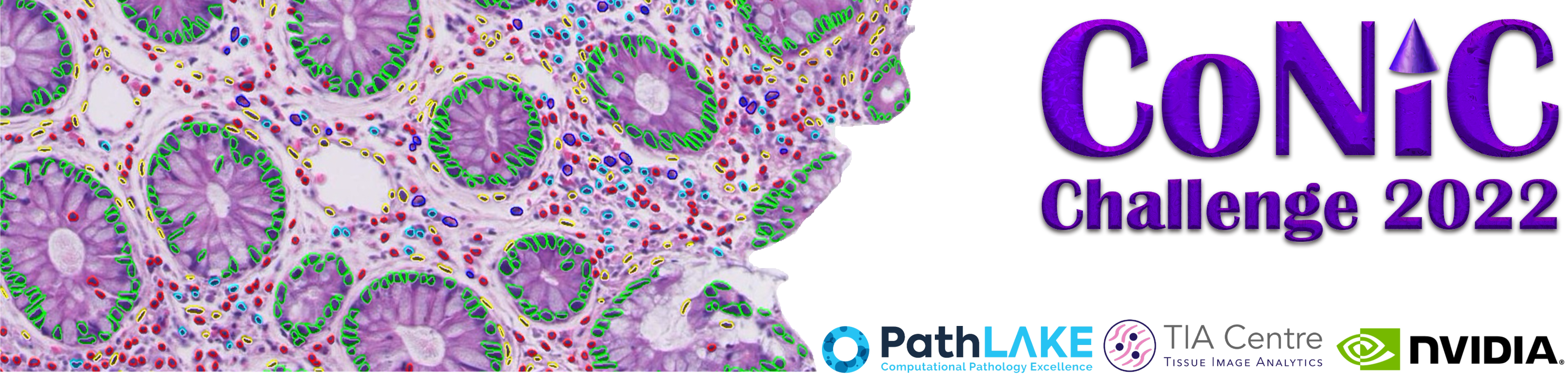
CoNIC: Colon Nuclei Identification and Counting Challenge¶
CoNIC: Colon Nuclei Identification and Counting Challenge¶
 About¶
About¶
Nuclear segmentation, classification and quantification within Haematoxylin & Eosin stained histology images enables the extraction of interpretable cell-based features that can be used in downstream explainable models in computational pathology (CPath). To help drive forward research and innovation for automatic nuclei recognition in CPath, we organise the Colon Nuclei Identification and Counting (CoNIC) Challenge. The challenge requires researchers to develop algorithms that perform segmentation, classification and counting of 6 different types of nuclei within the current largest known publicly available nuclei-level dataset in CPath, containing around half a million labelled nuclei.
To access code and example notebooks relating to the challenge, take a look at our GitHub page.
To access the code that we used for training the HoVer-Net baseline, we made a take a look at this link. Here, we made a new branch in the original HoVer-Net repository.
Take a look at our preprint for detailed information on the challenge.
NEWS: We are now accepting post-challenge submissions for both tasks! Refer to the final video in the submission tab for further details.¶
 Tasks¶
Tasks¶
This challenge consists of two separate tasks:
 Task 1: Nuclear segmentation and classification¶
Task 1: Nuclear segmentation and classification¶
The first task requires participants to segment nuclei within the tissue, while also classifying each nucleus into one of the following categories: epithelial, lymphocyte, plasma, eosinophil, neutrophil or connective tissue.
 Task 2: Prediction of cellular composition¶
Task 2: Prediction of cellular composition¶
For the second task, we ask participants to predict how many nuclei of each class are present in each input image.
The output of Task 1 can be directly used to perform Task 2, but these can be treated as independent tasks. Therefore, if it is preferred, prediction of cellular composition can be treated as a stand alone regression task.
 Challenge Format¶
Challenge Format¶
There are three main phases to this challenge:
- Discovery phase (Training/Validation)
- Preliminary test phase
- Final test phase
The CoNIC challenge starts with the discovery phase where we share the training data, challenge goals, and the evaluation code. Participants should start experimenting with the dataset and train/validate their model on it.
All challenge submissions are in the form of a docker container, which means that participants should submit their method to be evaluated on the test sets. In other words, only the training set will be released during the challenge and participants will not have access to any part of the test set.
The test will be done in two phases. The first phase, the preliminary test, will give participants a chance to work on their submissions for two weeks, get familiar with the submission workflow, improve their code if needed, and make sure their method works fine in the challenge evaluation pipeline. We will release a template docker structure and tutorials on how participants should package their code using the template and submit it to the challenge website. During the preliminary test phase, participants (teams) can have one submission per day. Note, the goal of this phase is to check your algorithm is working as expected. Right after the preliminary test phase, the final test phase begins where each team should submit their final method as a docker container, like before. Each final submission should be accompanied by a short manuscript describing technical details of the submitted method. If this is not done, then the submission will be disqualified. In the final test phase, each team has only one chance of submission per task, unless they are submitting two (or more) methods that are significantly different from each other.
 Important Dates¶
Important Dates¶
20th November 2021: Release of training data and evaluation code13th Feb 2022: Preliminary test set submission opens15th Feb 2022: Deadline for registration and team formation (mandatory for submission)28th Feb 2022: Deadline for 2 page method description2nd March 2022: Final test set submission opens8th March 2022: Notification of invitation for 4 page technical paper submission9th March 2022: Final test set submission closes20th March 2022: Deadline for submission of 4 page paper28th March 2022: CoNIC workshop at ISBI conference- 15th July 2022: Post-challenge submission opens

Check where you are in the challenge timeline.
 Registration¶
Registration¶
Individuals or team members interested in participating in this challenge should carefully study the challenge rules and then follow these steps:
- Register on the grand-challenge website via this link.
- Sign in to your account and revisit the challenge webpage.
- Click on the green "Join" bottom on the top right corner of the webpage.
After completing these three steps, you will be forwarded to the challenge registration page where you will need to submit your request. Once your participation request has been accepted, you will have access to the data download and submission pages.
*Please note that by participating in this challenge you are agreeing to all its rules and policies.
 Publication and Prizes¶
Publication and Prizes¶
Following the challenge, we will submit an overview of the results along with the described methods to a top tier medical image analysis journal. Participants will also get a chance to submit their work as part of the ISBI conference proceedings. Click here for more information.
The following prizes will be awarded for the challenge 🏆:
For each task:
1st place) \$500 + NVIDIA RTX 3070 GPU 🥇
2nd place) \$300 + NVIDIA Jetson Nano 🥈
3rd place) \$200 + NVIDIA Jetson Nano🥉
Note, to be eligible for prizes, algorithms must be made available (see rules).
 Citation¶
Citation¶
If you are comparing against any of the methods within the challenge or using our dataset, you must cite:
-
Graham, Simon, et al. "CoNIC Challenge: Pushing the frontiers of nuclear detection, segmentation, classification and counting." Medical image analysis 92 (2024): 103047.
-
Graham, Simon, et al. "Lizard: A Large-Scale Dataset for Colonic Nuclear Instance Segmentation and Classification." Proceedings of the IEEE/CVF International Conference on Computer Vision. 2021.